
The Zappo scheme colors the sequences according to their biophysical properties: The sequences in this example are too short and simple to see the impact of iterations so we are going to use the default settings.Īlign these sequences with ClustalOmega using default parameters. That's why the default setting is to not use the iterations. Each iteration will add 1-3 times it costs to make the alignment without iterations. Iterations come at a cost, they increase the run time. The number of times it refines the alignment is defined by the Max number of HMM iterations parameter.

#Bioedit make a tree download
You can download the sequences in fasta format. We are first going to use different tools for the toy example below:
#Bioedit make a tree manual
Some MSA tools allow manual editing of alignments, which will be discussed in the next exercise. Let's discover how some of these programs behave, and which suit your needs best! As always, EBI provides a nice collection of web-based alignment tools but you can also make MSAs in Ugene. Fortunately, you can also find tools to convert MSA formats.
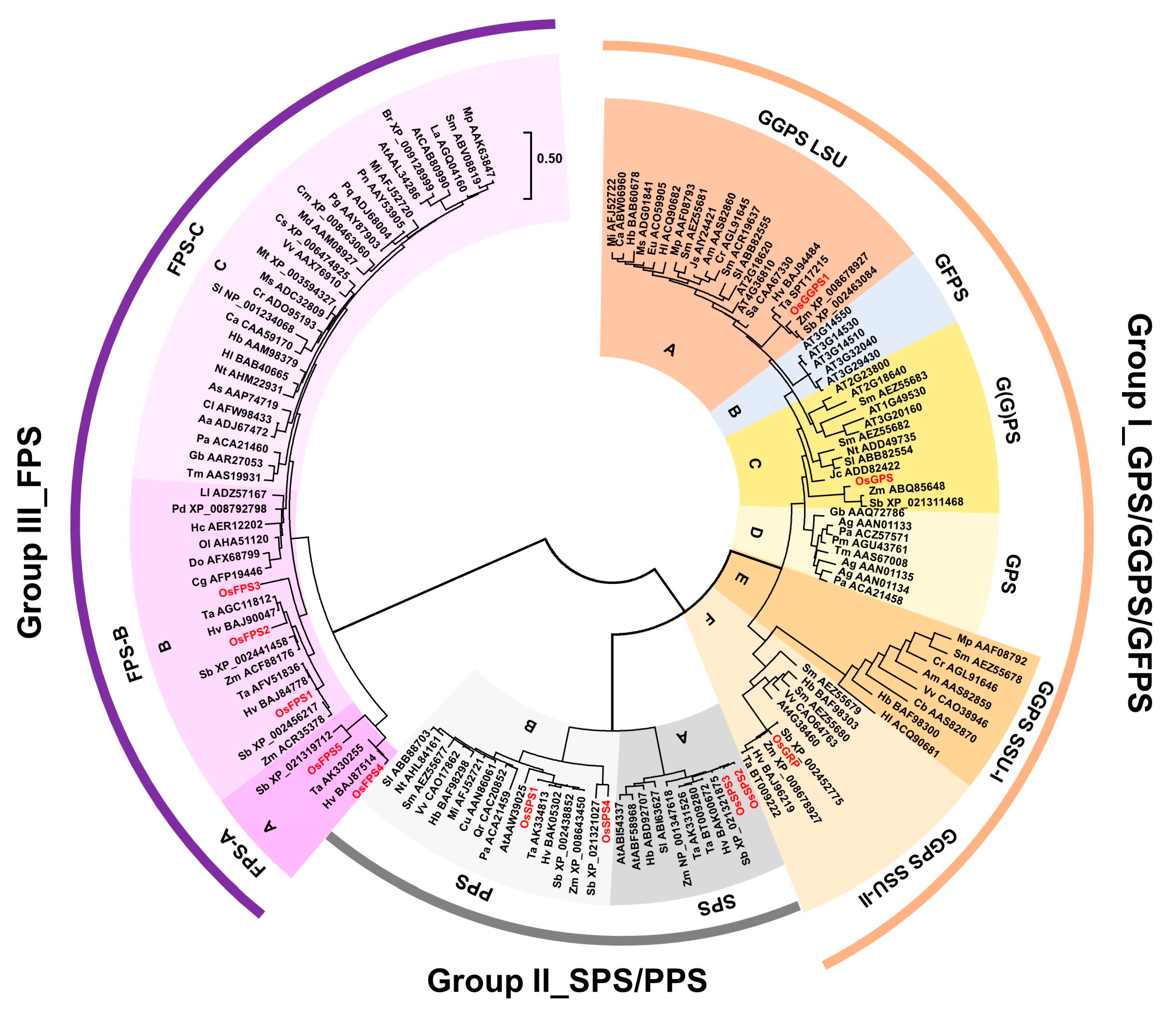
Several multiple sequence algorithms exist, each with their own program and format.


 0 kommentar(er)
0 kommentar(er)
